In the previous post, I went over some basic stuff for using the sqlite database with the sqlite3 package for Python. Manipulating data with Python is so easy and time-saving, it really is a good idea.
This time I'm going over a common operation for data-wrangling that I found very useful. I will be:
- reading a CSV data file source (which has been exported or transformed from somewhere else)
- parsing the file to guess information about its columns and datatypes
- creating a sqlite database table automatically based on those guesses
- reading a batch of lines at a time from the CSV file, then
- inserting the batch of information into the new database table
After doing whatever I like to my new handy information in the database, I will be:
- fetching a batch of records at a time from the database
- writing the batch of records into a new CSV output file.
The point of that is that I want a better, wrangled final CSV containing just what I want, for purposes like training with it for machine learning.
To do this, I have some Python code files and my sample data CSV file. The scripts below will be just be run on the command line. You might also see some screenshots from SQLiteStudio (if you haven't got it, it's great! A GUI tool for sqlite).
First of all, I have a
CSVHandler class to do my dirty work for me. This
will read from or write to a CSV file. It will need methods to (a) make intelligent guesses on column data types, (b) read from a group of lines in the file at a time, and (c) append a set of lines to a CSV file at a time.
import csv
"""
A class for dealing with CSV files for
(a) reading from a CSV file
(b) writing to a CSV file
"""
class CSVHandler:
fieldTypeString = "TEXT"
fieldTypeInt = "INTEGER"
fieldTypeFloat = "REAL"
def __init__(self, inputFilePath="", outputFilePath=""):
"""Constructor:
:param inputFilePath: a string to specify the path to a CSV file to read from.
:param outputFilePath: a string to specify the path to a CSV file to write to.
"""
self.inputFilePath = inputFilePath
self.outputFilePath = outputFilePath
def isFloat(self, s):
"""
:param s: a string for a value.
:rtype: bool
"""
try:
float(str(s))
return True
except ValueError:
return False
def isInt(self, s):
"""
:param s: a string for a value.
:rtype: bool
"""
try:
val = int(str(s))
return True
except ValueError:
return False
def getColumnsInfo(self):
"""
Attempts to make intelligent guesses to what data types for SQLite
the records in a CSV file match to.
:rtype: dict
"""
sampleRows = self.readPartial(0, 24)
firstRow = sampleRows[0]
keys = [x for x in firstRow.keys()]
# Do some checking for each column's values to see which data type it
# *doesn't* match. Possible combinations:
# {
# 't_string': {'INTEGER': True, 'REAL': True},
# 't_float': {'INTEGER': True, 'REAL': False}
# 't_int': {'INTEGER': False, 'REAL': False},
# }
errorChecks = {}
for key in keys:
errorChecks[key] = {
self.fieldTypeInt: False,
self.fieldTypeFloat: False
}
for row in sampleRows:
for key, value in row.items():
if self.isInt(value) == False:
errorChecks[key][self.fieldTypeInt] = True
if self.isFloat(value) == False:
errorChecks[key][self.fieldTypeFloat] = True
info = {}
for key in keys:
if errorChecks[key][self.fieldTypeInt] == True and \
errorChecks[key][self.fieldTypeFloat] == True:
info[key] = self.fieldTypeString # set a default data type
elif errorChecks[key][self.fieldTypeInt] == True:
info[key] = self.fieldTypeFloat
else:
info[key] = self.fieldTypeInt
return info
def getTotalRows(self):
"""
Returns the total number of lines found in a CSV file.
:rtype: int
"""
with open(self.inputFilePath) as csvfile:
csvReader = csv.reader(csvfile)
for row in csvReader:
pass
return csvReader.line_num
# Alternative counter:
# gen = (row for row in csvReader)
# return len(list(gen))
def readPartial(self, limit=-1, offset=0):
"""
Reads a set of lines from a CSV file.
:param limit: a number of the max limit number of rows to read.
:param offset: a number for the start index row to start reading at.
:rtype: list
"""
with open(self.inputFilePath) as csvfile:
csvReader = csv.DictReader(csvfile)
for i in range(offset):
next(csvReader)
rows = []
count = 0
for row in csvReader:
rows.append(row)
count += 1
if count == limit:
break
return rows
def writePartial(self, fieldNames, rows, isInitial=True):
"""
Writes/appends a set of lines to a CSV file.
:param fieldNames: a list of the column names for the CSV file header row.
:param rows: info for a set of rows to be inserted (a list of dicts)
:param isInitial: whether this will be the first row in the file (or not)
"""
with open(self.outputFilePath, 'a') as csvfile:
writer = csv.DictWriter(csvfile, fieldnames=fieldNames)
if isInitial == True:
writer.writeheader()
for row in rows:
writer.writerow(row)
Next, I'll need a class called DBConnector to deal with a SQLite database and one particular table. It will need to:
(a) Create a table based on info I give it,
(b) fetch the field names and total number of records in the table, and
(c) insert a batch of 'X' number of records
(d) fetch a batch of 'X' number of records
"""
An object for connecting to a SQLite database.
"""
import sqlite3
class DBConnector:
def __init__(self, dbLocation):
"""Constructor:
:param dbLocation: a string to specify the path to the database file.
"""
self.connection = sqlite3.connect(dbLocation)
self.connection.row_factory = sqlite3.Row
self.cursor = self.connection.cursor()
def createTable(self, tableName, columnsInfo):
"""
Creates a new table in the database (if it doesn't exist yet).
:param tableName: the new table's name.
:param columnsInfo: a dict with info about table fields
:rtype: dict
"""
result = {"data": [], "errors": ""}
# Build a list of column name / field type statements
params = []
for columnName, affinity in columnsInfo.items():
params.append(columnName + " " + affinity)
q = """create table {} (
id integer primary key autoincrement not null,
{}
)""".format(tableName, ",\n".join(params))
try:
self.cursor.execute(q, [])
self.connection.commit()
result["data"].append("success")
except Exception as e:
result["errors"] = str(e)
return result
def getTotalRows(self, tableName):
"""
:param tableName: the new table's name.
Returns the count of records in a query result.
:rtype: dict
"""
result = {"data": 0, "errors": ""}
q = "select count(id) as total from " + tableName
try:
res = self.cursor.execute(q, ()).fetchone()
if res is not None:
result["data"] = res[0]
except Exception as e:
result["errors"] = str(e)
return result
def getFieldNames(self, tableName):
"""
:param tableName: the new table's name.
Returns the list of field names from a database table.
:rtype: dict
"""
result = {"data": [], "errors": ""}
q = "SELECT name FROM PRAGMA_TABLE_INFO('" + tableName + "')"
try:
res = self.cursor.execute(q, ()).fetchall()
for fieldRow in res:
result["data"].append(fieldRow[0])
except Exception as e:
result["errors"] = str(e)
return result
def doBatchInsert(self, tableName, fieldNames, rowsBatch):
"""
:param tableName: the new table's name.
:param fieldNames: the list of fields
:param rowsBatch: info for a batch of records (a list of dictionaries.)
:rtype: dict
"""
allParams = []
recordVals = []
# Build a query with bind parameters (?) where values should go.
for row in rowsBatch:
params = []
for fieldName in fieldNames:
params.append("?") # bind parameter
allParams.append(row[fieldName])
recordVals.append("(" + ", ".join(params) + ")")
q = "insert into {} ({}) values {}".format(tableName,
", ".join(fieldNames),
",\n".join(recordVals)
)
return self.insert(q, allParams)
def selectMany(self, q, params, limit=500, offset=None):
"""
Runs a select query.
:param q: the database query string.
:param params: a list of any bound parameters to be inserted into the query string.
:return result: a set of info from running the query
:rtype: dict
"""
result = {"data": [], "errors": ""}
q += " limit " + str(limit)
if offset is not None:
q += " offset " + str(offset)
try:
res = self.cursor.execute(q, params).fetchall()
result["data"] = [dict(row) for row in res]
except Exception as e:
result["errors"] = str(e)
return result
Here is my first script to operate using those classes defined earlier 'insert_records_script.py'. It will (a) read an input CSV file to get info about its columns, (b) create a table in the database and (c) read a batch of CSV lines at a time, then (d) insert each batch at a time into the database.
from lib.DBConnector import DBConnector
from lib.CSVHandler import CSVHandler
"""
A script for inserting records from a source CSV file into a SQLite database
table.
"""
if __name__ == "__main__":
csvFilePath = "./data/serious-injury-outcome-indicators-2000-19-csv.csv"
dbFileLocation = "/home/scott/databases/sqlite/test_db1.db"
newTableName = "test_tbl"
batchSizeForInserting = 100
# Read a source CSV file to extract some info about its number of rows and
# typical data found in the columns.
csvHandler = CSVHandler(csvFilePath)
columnsInfo = csvHandler.getColumnsInfo()
totalRecords = csvHandler.getTotalRows()
# Connect to the Database
conn = DBConnector(dbFileLocation)
# Create a table in the DB with field types based on intelligent guesses
# from reading the CSV source data file.
result = conn.createTable(newTableName, columnsInfo)
# Insert a batch of records at a time, until all the rows from the CSV
# file we read have been used up.
fieldNames = columnsInfo.keys()
for offset in range(0, totalRecords, batchSizeForInserting):
print("Insert records at offset: " + str(offset))
rowsBatch = csvHandler.readPartial(batchSizeForInserting, offset)
result = conn.doBatchInsert(newTableName, fieldNames, rowsBatch)
print(result)
This is me running the "insert" script:
Now I have my info in the sqlite3 database, sweet! It's so much easier to do common operations in a relational database than a clunky, inefficient spreadsheet.
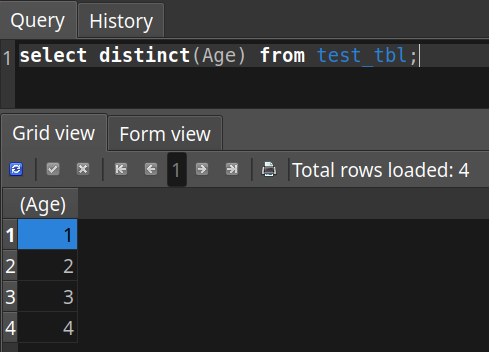
Now, as an example of something I want do for wrangling, there is a column in the table, 'Ages', that has text info. This info could be used to represent categories of data. To make operations on it later easier for things like machine learning, I want to transform the text values into numerical values so they can be counted more easily by applications like tensorflow.
So I run some update commands, and now the values are numerical. I'm happy. I might also think about columns I won't need next time.
update test_tbl set Age = '1' where Age = 'All ages';
update test_tbl set Age = '2' where Age = '0-74 years';
update test_tbl set Age = '3' where Age = '75+ years';
update test_tbl set Age = '4' where Age = '0-14 years';
I want to now export the data out of the SQLite database table and into a new CSV output file. My next script 'retrieve_records_script.py' will:
(a) Find out what the table columns are and the number of records I'm dealing with, and (b) fetch a batch of records at a time from the database, then (c) write each batch of records to an output CSV file.
from lib.DBConnector import DBConnector
from lib.CSVHandler import CSVHandler
"""
A script for fetching records from a SQLite database, and writing them into
a new output CSV file.
"""
if __name__ == "__main__":
csvFilePath = "./data/new_serious-injury-outcome-indicators.csv"
dbFileLocation = "/home/scott/databases/sqlite/test_db1.db"
newTableName = "test_tbl"
batchSizeForFetching = 100
# Connect to the Database
conn = DBConnector(dbFileLocation)
totalRecords = conn.getTotalRows(newTableName)["data"]
fieldNames = conn.getFieldNames(newTableName)["data"]
csvHandler = CSVHandler(inputFilePath="", outputFilePath=csvFilePath)
# Fetch a batch of records at a time from the database, then write the
# batch to an output CSV file.
batchFetchQuery = "select * from " + newTableName
params = ()
for offset in range(0, totalRecords, batchSizeForFetching):
print("Fetch records at offset: " + str(offset))
# Select query
result = conn.selectMany(batchFetchQuery, params, batchSizeForFetching, offset)
# Write the batch of records to the output CSV file
print("Writing " + str(len(result["data"])) + " rows to the CSV output file")
isInitial = False
if offset == 0:
isInitial = True
csvHandler.writePartial(fieldNames, result["data"], isInitial)
print("CSV output file written: " + csvFilePath)
Here's me running the 'retrieve' script.
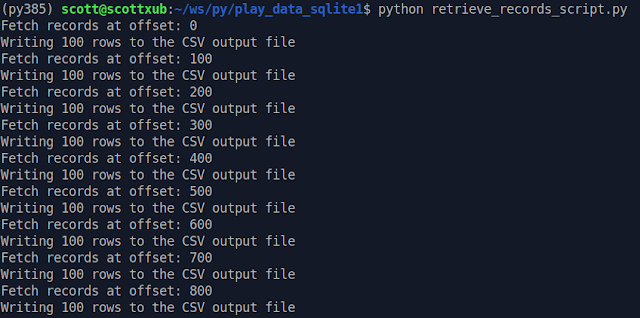
And here's my new CSV output file. All's well that end's well! I have a file that an application can suck in my wrangled data easily, and I didn't nearly crash my computer waiting for Excel to do unwieldy data operations.